Welcome to the Larrondo Lab
|
The Larrondo Lab is interested in different aspects of gene expression. In particular, we study the molecular mechanisms underlying biological oscillators, assessing the impact that circadian clocks have on physiology and in host-pathogen interactions. We use various fungal organisms as model systems to study these complex biological processes: the filamentous fungus Neurospora crassa, the phytopathogen Botrytis cinerea, the biocontroller Trichoderma atroviride and the yeast Saccharomyces cerevisiae.
Our studies employ functional genomics strategies, as well as the use of real-time reporters. Lately, we have began adopting optogenetics and synthetic biology approaches to attempt to design synthetic circuits capable of rhythmicity or predictable behaviors, with the goal of studying the design principles underlying these systems. We are additionally interested in applying some of this knowledge to tackle different biotechnological challenges [Read More]. |
|
Our laboratory is in the Department of Molecular Genetics and Microbiology at the School of Biological Sciences, Pontificia Universidad Catolica de Chile, and part of the iBio
Projects
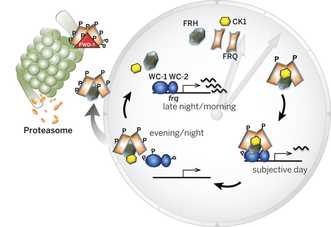
Mechanisms of the Neurospora circadian clockWe are interested in the mechanisms underlying the workings of the Neurospora pacemaker, as well as in the different ways in which it regulates rhythmic gene expression [Read More].
|
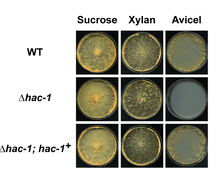
Plant cell wall deconstructionWe attempt to characterize the molecular mechanisms through which fungi degrade plant biomass [Read More].
|
FUNGAL SYNTHETIC BIOLOGY
Optogenetics
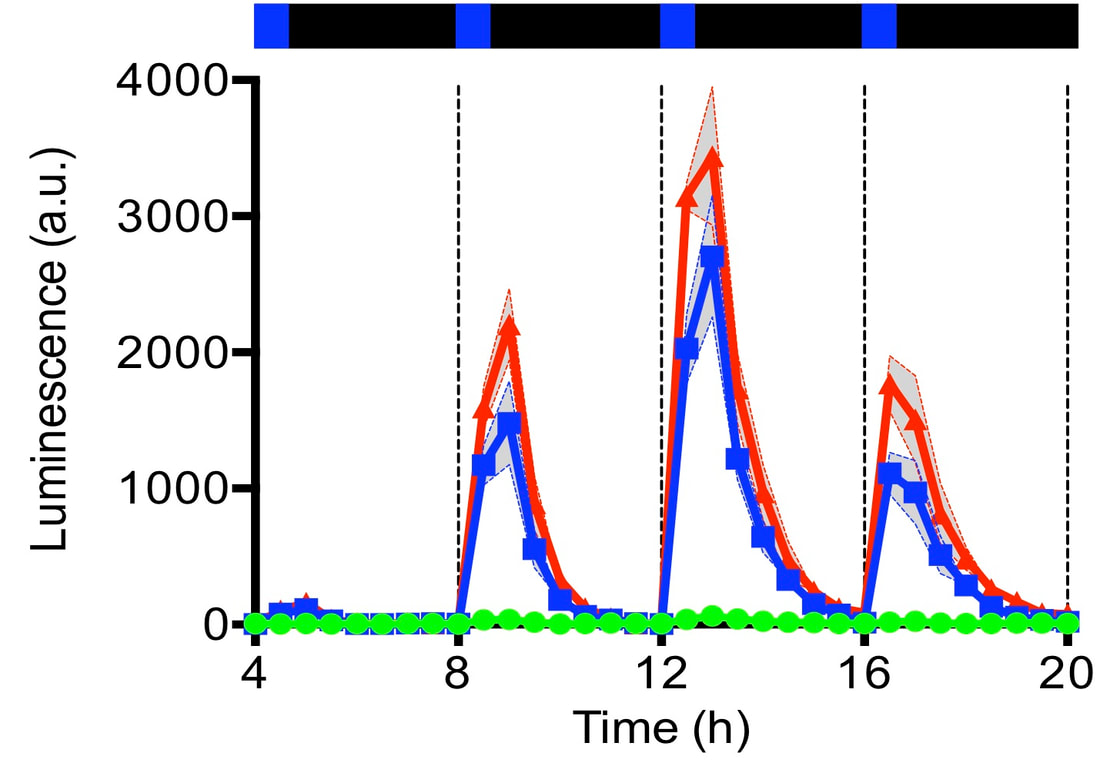
We study how light responses can be utilized to control gene expression, modifying existing programs or building artificial optogenetic switches [Read More].
Oscillators
|
current Funding sources
Copyright © 2016. All rights reserved. Larrondo Lab.